Hypoxylon sp. 14171
| Strain |
14171 |
| Classification |
Xylariales, Hypoxylaceae, Hypoxylon |
| Culture collection |
processing |
| Detection frequency |
Low |
| Figure |
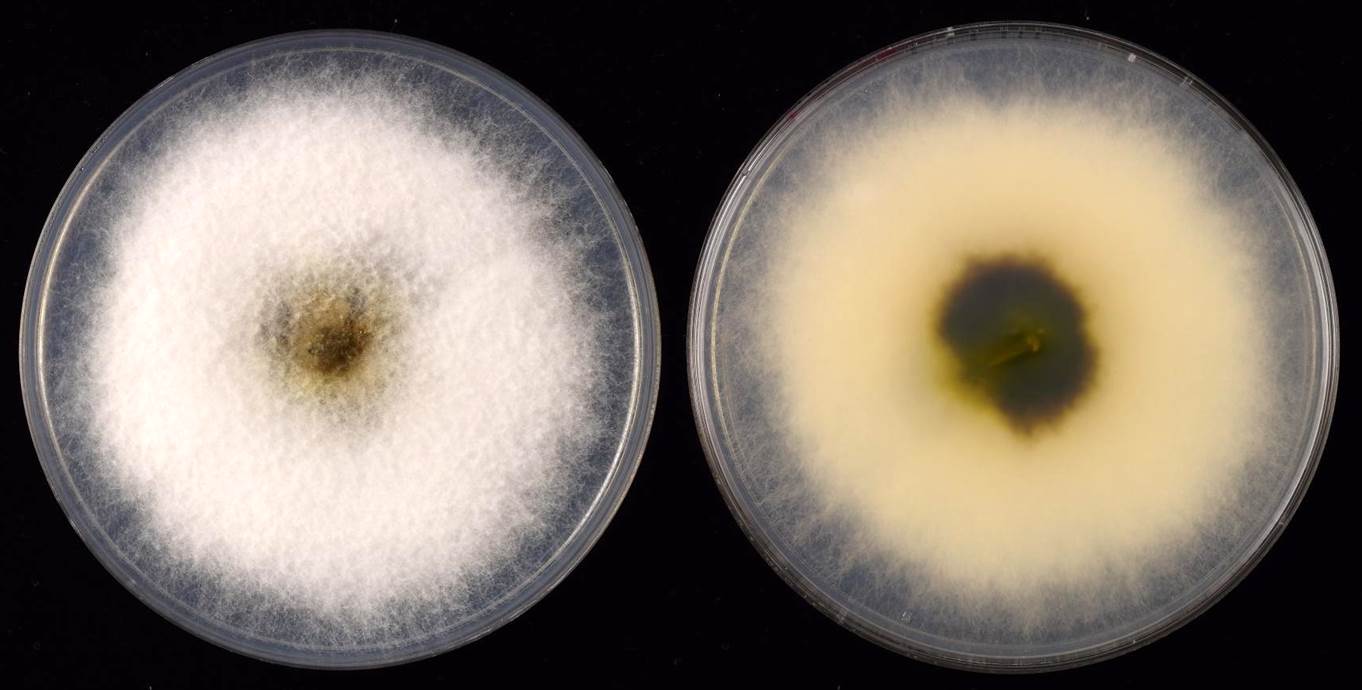 Fig. 1 5-day-old colony on PDA Fig. 1 5-day-old colony on PDA
 Fig. 2 Hypoxylon sp. 14171. a–c. Conidiophores, conidiogenous cells and conidia. d. Conidiogenous cells. e. Conidia (Bars= 5 μm, unless otherwise specified) Fig. 2 Hypoxylon sp. 14171. a–c. Conidiophores, conidiogenous cells and conidia. d. Conidiogenous cells. e. Conidia (Bars= 5 μm, unless otherwise specified) |
| Colonies |
Colonies on PDA attaining >90-mm at 25 °C after 5 days, cottony, initially white, turning dark olivaceous brown from the center with age, margins entire, reverse similar, completely turning black in older culture. |
| Conidiophores |
Conidiophores erect, brown, rough-walled, often with dark brown exudates, paler toward the apex, with several lateral branches, terminating in 1–6 verticillately arranged conidiogenous cells, 2–3.5 µm wide, up to 300 µm long. |
| Conidiogenous cells |
Conidiogenous cells cylindrical, pale brown, mostly terminal, rarely intercalary, slightly denticulate near the conidiogenous loci, (6–) 9–15(–19) × 2–3 µm. |
| Conidia |
Conidia one-celled, smooth, subhyaline, obovoid, with a slightly truncated base, 2.5–3.5 × 1.5–2.5 µm. |
| Note |
This fungus produced asxual conidia of Nodulisporium-state very well on PDA medium. Base on the phylogenetic relationships inferring from ITS region, this strain is clustered within H3 clade defined by Wendt et al. (2018). The closest species of this strain is Hypoxylon lenormandii strain CBS 119003 (90.39%; KC968943). The identity of this strain is still uncertain. |
| Pathogenicity |
Unknown |
| Specimens examined |
Taiwan, Taichung City, rice grains (cultivar Taichung 194), Dec 2014, Jie-Hao Ou, 14171 |
| ITS |
AAAACTCCAAACCCTTTGTGAACCTTACCGTCGTTGCCTCGGCGTGAGCTACGGCTACCCGGGAGCTACCCTGGAGCTACCCTAGAGTTACCCTATAGCTACCCTGCAGCTACCCTATACTTACCCTATAGCTACCCTGCAGCTACCCTATAGTTAGTTACCCTAGAGTTACCCTGGAGCTACCCTGTAGCCGGCTTATGGCCCGCCGAAGGACAGCTAAACTCTTGTTTTTTACTACTGTTTCTCTGAATTACAAACTGAAATAAGTTAAAACTTTCAACAACGGATCTCTTGGTTCTGGCATCGATGAAGAACGCAGCGAAATGCGATAAGTAATGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCGCCCATTAGTATTCTAGTGGGCATGCCTATTCGAGCGTCATTTCGACCCCTAAGCCCCTGTTGCTTAGCGTTGGGAATCTACAGCGTAGTTCCTCAAAATTAGTGGCGGAGTCAGGGTACACTCTCAGCGTAGTAATTTCTCTCGCTCGTGTGGTGGCCTTGGCTGCTAGCCGTTAAAACCCCTATAATTTCTAGTGGTTGACCTCGGATTAGGTAGGAATACCCGCTGAACTTAA |
 Fig. 1 5-day-old colony on PDA
Fig. 1 5-day-old colony on PDA
 Fig. 2 Hypoxylon sp. 14171. a–c. Conidiophores, conidiogenous cells and conidia. d. Conidiogenous cells. e. Conidia (Bars= 5 μm, unless otherwise specified)
Fig. 2 Hypoxylon sp. 14171. a–c. Conidiophores, conidiogenous cells and conidia. d. Conidiogenous cells. e. Conidia (Bars= 5 μm, unless otherwise specified)