Curvularia tuberculata
| Authors |
B.L. Jain 1962 |
| Strain |
12005 |
| Classification |
Pleosporales, Pleosporaceae, Curvularia |
| Culture collection |
BCRC FU30161 |
| Detection frequency |
Low |
| Accession number |
LC494371 |
| Figure |
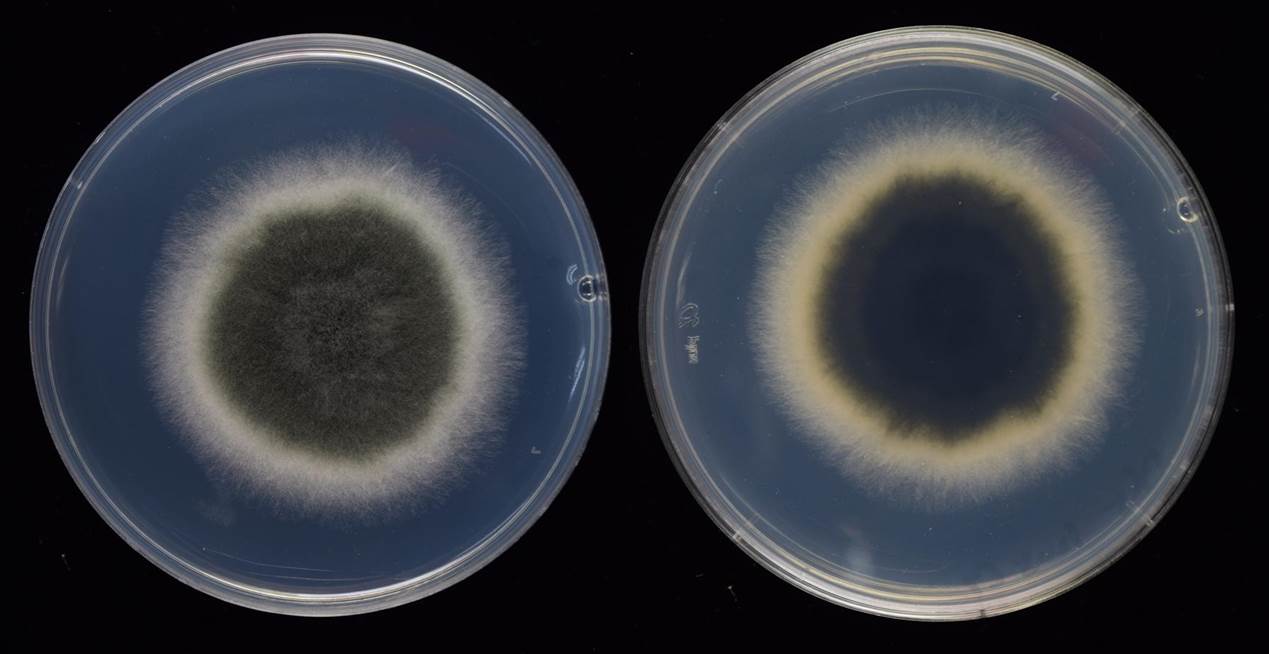 Fig. 1 5-day-old colony on PDA Fig. 1 5-day-old colony on PDA  Fig. 2 Curvularia tuberculata. a–c. Conidiophores and conidia. d, e. Conidia (Bars= 5 μm, unless otherwise specified) Fig. 2 Curvularia tuberculata. a–c. Conidiophores and conidia. d, e. Conidia (Bars= 5 μm, unless otherwise specified) |
| Colonies |
Colonies on PDA attaining 54-, 76-, 23-mm at 24 °C, 28 °C and 37 °C after 5 days, margins entire, floccose, reverse dark olivaceous to black with white margins, slightly zonated. |
| Conidiophores |
Conidiophores macronematous, light brown, flexuous, septate, simple or branched, up to 200 µm, 3–3.5 µm wide. |
| Conidiogenous cells |
Conidiogenous cells integrated, terminal or intercalary, geniculate, sympodial, echinulate near the conidiogenous loci. |
| Conidia |
Conidia straight, rarely curved, cylindrical, ellipsoidal or clavate, tuberculate, with slightly darkened and thickened hilum, olivaceous brown, 2-4 septate(mostly 3), 25–50 × 12–20 µm. |
| Note |
The species was commonly reported on diverse host plants as being saprobic or pathogenic, occasionally on humans as opportunistically pathogenic. Strain 12005 shares 100% identity in ITS region with the iso-type strain of Curvularia tuberculata (CBS 146.63, JX256433). |
| Pathogenicity |
This species is often found on rice seeds and may be associated with rice discoloration (Misra & Dharam, 1988). This species was reported to cause leaf spot in Pakistan (Majeed et al., 2016). |
| Specimens examined |
Taiwan, Tainan City, rice grains (cultivar Tainan 11), Sep 2012, Jie-Hao Ou, 12005 |
| ITS |
AGGAAGTAAAAGTCGTAACAAGGTCTCCGTAGGTGAACCTGCGGAGGGATCATTACACAAAAAAATATGAAGGCTGACTGCAGCTGACCTACATGGCCAGTTTGCTCGAGGCTGAATTATTTTTTCACCCATGTCTTTTGCGCACTTGTTGTTTCCTGGGCGGGTTCGCCCGCCACCAGGACCACACCATAAACCTTTTTATCTGTAGTTGCAATCAGCGTCAGTAACAAGTAAATAATCATTTACAACTTTCAACAACGGATCTCTTGGTTCTGGCATCGATGAAGAACGCAGCGAAATGCGATACGTAGTGTGAATTGCAGAATTCAGTGAATCATCGAATCTTTGAACGCACATTGCGCCCTTTGGTATTCCAAAGGGCATGCCTGTTCGAGCGTCATTTGTACCCTCAAGCTTTGCTTGGTGTTGGGCGTTTTTGTCTTTGGCTACAGCCCGAAGACTCGCCTTAAAACGATTGGCAGCCGGCCTACTGGTTTCGCAGCGCAGCACATTTTTGCGCTTGCAATCAGCAAAAGAGGTTGGCGATCCATCAAGT |
 Fig. 1 5-day-old colony on PDA
Fig. 1 5-day-old colony on PDA  Fig. 2 Curvularia tuberculata. a–c. Conidiophores and conidia. d, e. Conidia (Bars= 5 μm, unless otherwise specified)
Fig. 2 Curvularia tuberculata. a–c. Conidiophores and conidia. d, e. Conidia (Bars= 5 μm, unless otherwise specified)