| Authors | Link 1809 |
|---|---|
| Strain | 13049 |
| Classification | Eurotiales, Aspergillaceae, Aspergillus |
| Culture collection | BCRC FU30219 |
| Detection frequency | Medium |
| Accession number | LC494361 |
| Figure | 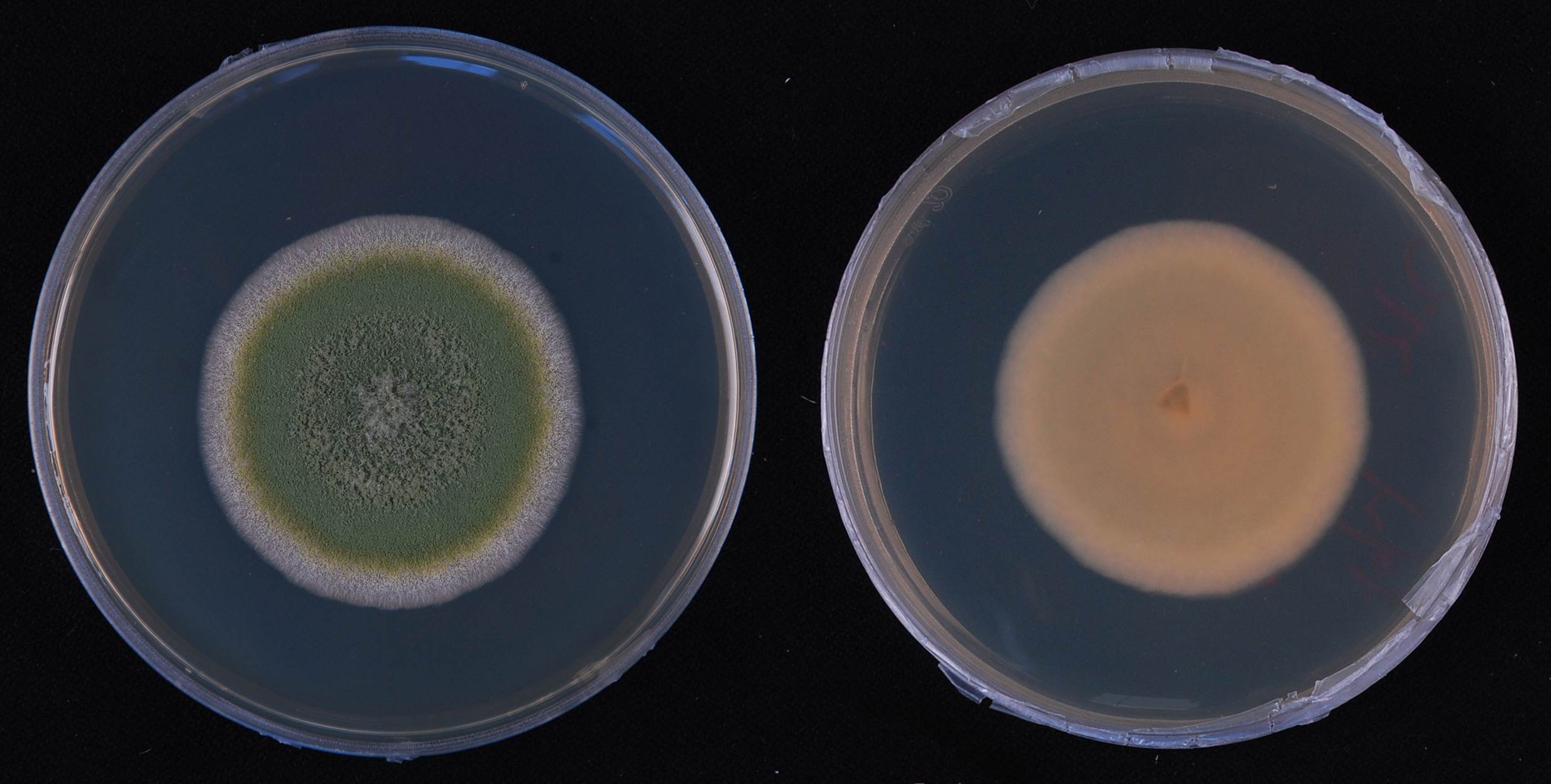 Fig. 1 5-day-old colony on PDA Fig. 1 5-day-old colony on PDA  Fig. 2 Aspergillus flavus. a–d. Conidiophores and conidia. e. Conidia (Bars= 5 μm, unless otherwise specified) Fig. 2 Aspergillus flavus. a–d. Conidiophores and conidia. e. Conidia (Bars= 5 μm, unless otherwise specified) |
| Colonies | Colonies grow well on PDA at 37°C, attaining 62, 80, 49 mm at 24 °C, 28 °C and 37 °C after 5 days, margins entire, yellow-green, floccose, zonated, reverse white to yellowish. |
| Conidiophores | Conidiophores, straight, hyaline, rough-walled, with swollen apex, up to 1 mm long, 8–15 µm wide. |
| Conidiogenous cells | Conidiogenous cells phialidic, ampulliform, hyaline to pale brown, mostly uniseriate, 8–10 × 3.5–5 µm. |
| Conidia | Conidia hyaline, golden brown,when mature, radial orientation, catenate, often more than 70 conidia in chain, globose to ellipsoidal, smooth, 3–6 µm in diameter. |
| Note | During this study, this species was found frequentlyon stored rice seeds and occasionally on fresh harvested rice seeds. |
| Pathogenicity | This species is considered as a saprophytic fungus and only occasionally causes grain discoloration in storage (Ou, 1985). Some strains of this species have the ability to produce aflatoxin. According to different investigations, about 50-70% of the strains have the ability to produce aflatoxin (Chhotaray et al., 2015; Guimarães et al., 2010). |
| Specimens examined | Taiwan, Chiayi County, rice grains (cultivar Tainan 11), Sep 2013, Jie-Hao Ou, 13049 Taiwan, Tainan City, rice grains (cultivar Tainan 11), Jul 2014, Jie-Hao Ou, 14081 Taiwan, Taichung City, rice grains (cultivar Taichung 65), Feb 2015, Jie-Hao Ou, 15010 |
| ITS | GTGTAGGGTTCCTAGCGAGCCCAACCTCCCACCCGTGTTTACTGTACCTTAGTTGCTTCGGCGGGCCCGCCATTCATGGCCGCCGGGGGCTCTCAGCCCCGGGCCCGCGCCCGCCGGAGACACCACGAACTCTGTCTGATCTAGTGAAGTCTGAGTTGATTGTATCGCAATCAGTTAAAACTTTCAACAATGGATCTCTTGGTTCCGGCATCGATGAAGAACGCAGCGAAATGCGATAACTAGTGTGAATTGCAGAATTCCGTGAATCATCGAGTCTTTGAACGCACATTGCGCCCCCTGGTATTCCGGGGGGCATGCCTGTCCGAGCGTCATTGCTGCCCATCAAGCACGGCTTGTGTGTTGGGTCGTCGTCCCCTCTCCGGGGGGGACGGGCCCCAAAGGCAGCGGCGGCACCGCGTCCGATCCTCGAGCGTATGGGGCTTTGTCACCCGCTCTGTAGGCCCGGCCGGCGCTTGCCGAACGCAAATCAATCTTTTTCCAGGTTGACCTCGGATCAGGTAGGGATACCCGCTGAACTTAA |